What'sNEW September–October 2010
 31 October 2010 31 October 2010
A layer of organic compounds may give Kuiper-Belt objects their colors. This suggestion comes from physicist John Cooper of NASA's Goddard Space Flight Center, at the October meeting of the American Astronomical Society in Pasadena CA. The Kuiper Belt is the orbital zone where more than 1,000 icy bodies, including Pluto, have been detected so far. Normally, sunlight cooks exposed organics into a tarry black crust, but the colors of these objects range from red to blue to white. However, if the organics are buried under a protective layer, with occasional and varying exposure, the colors can be explained. Of course, Cooper does not suggest that life made the organics, but we do!
 Kuiper Belt of Many Colors by NASA's Karen C. Fox, posted on redOrbit.com, 28 Oct 2010. Kuiper Belt of Many Colors by NASA's Karen C. Fox, posted on redOrbit.com, 28 Oct 2010.
 Icy Red Objects at Solar System's Edge May Point to Life's Building Blocks, Space.com, 28 Oct 2010. Icy Red Objects at Solar System's Edge May Point to Life's Building Blocks, Space.com, 28 Oct 2010.
 Comets: The Delivery System is the main related local webpage. Search for "Kuiper" for related articles. Comets: The Delivery System is the main related local webpage. Search for "Kuiper" for related articles.
 Thanks again, Jacob Navia. Thanks again, Jacob Navia.
 26 October 2010 26 October 2010
We're finding suites of genes that you would really never expect to find in viral life, but would expect to find in cellular organisms — marine virologist Curtis Suttle of the University of British Columbia
American, British and Canadian biologists have sequenced a newly discovered giant marine virus designated Cafeteria roenbergensis virus (CroV). It has a double-stranded DNA genome of ~730 kb, with 544 predicted protein-coding genes. Half of these have no match among sequenced genomes (see pie-chart), but many others have various apparent functions among higher life forms:
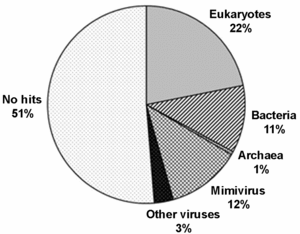 "The diverse coding potential of CroV includes predicted translation factors, DNA repair enzymes such as DNA mismatch repair protein MutS and two photolyases, multiple ubiquitin pathway components, four intein elements, and 22 tRNAs. Many genes including isoleucyl-tRNA synthetase, eIF-2γ, and an Elp3-like histone acetyltransferase are usually not found in viruses. We also discovered a 38-kb genomic region of putative bacterial origin, which encodes several predicted carbohydrate metabolizing enzymes, including an entire pathway for the biosynthesis of 3-deoxy-d-manno-octulosonate, a key component of the outer membrane in Gram-negative bacteria."
"The diverse coding potential of CroV includes predicted translation factors, DNA repair enzymes such as DNA mismatch repair protein MutS and two photolyases, multiple ubiquitin pathway components, four intein elements, and 22 tRNAs. Many genes including isoleucyl-tRNA synthetase, eIF-2γ, and an Elp3-like histone acetyltransferase are usually not found in viruses. We also discovered a 38-kb genomic region of putative bacterial origin, which encodes several predicted carbohydrate metabolizing enzymes, including an entire pathway for the biosynthesis of 3-deoxy-d-manno-octulosonate, a key component of the outer membrane in Gram-negative bacteria."
All of this is unsurprising if viruses are primary agents for horizontal gene transfer (HGT), and if HGT is essential for evolutionary progress, as we believe.
 Matthias G. Fischer et al., "Giant virus with a remarkable complement of genes infects marine zooplankton" [abstract], doi:10.1073/pnas.1007615107, Proc. Natl. Acad. Sci. USA, online 25 Oct 2010. Matthias G. Fischer et al., "Giant virus with a remarkable complement of genes infects marine zooplankton" [abstract], doi:10.1073/pnas.1007615107, Proc. Natl. Acad. Sci. USA, online 25 Oct 2010.
 Giant marine virus found by Jef Akst, The Scientist, 25 Oct 2010. Once thought not to exist in marine environments, scientists now realize that there are some 50 million viruses in every milliliter of seawater. Giant marine virus found by Jef Akst, The Scientist, 25 Oct 2010. Once thought not to exist in marine environments, scientists now realize that there are some 50 million viruses in every milliliter of seawater.
 Viruses and Other Gene Transfer Mechanisms is the main related local webpage — What'sNEW about HGT Viruses and Other Gene Transfer Mechanisms is the main related local webpage — What'sNEW about HGT  | |
 Thanks, Jacob Navia. Thanks, Jacob Navia.
 25 October 2010 25 October 2010
A compendium of articles from the online Journal of Cosmology, with two new ones, is available.
 The Biological Big Bang: Panspermia and Origins of Life, Chandra Wickramasinghe, ed., v11, Journal of Cosmology, 25 Oct 2010. The Biological Big Bang: Panspermia and Origins of Life, Chandra Wickramasinghe, ed., v11, Journal of Cosmology, 25 Oct 2010.
 25 October 2010 25 October 2010
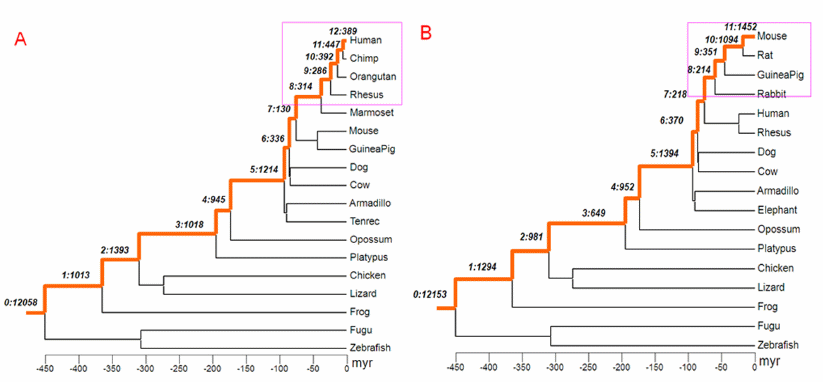
Genes are either very old, or they appear suddenly, without predecessors. This observation follows our long, ongoing study of the relevant scientific literature. The latest installment comes from an analysis of "male-biased" genes during mammalian evolution by biologists in Chicago and France. The researchers are curious to know "how the age of genes affects their chromosomal locations." We are curious about the provenance of newly acquired genetic programs, also indirectly illuminated in the report.
|
Phylogenetic tree of vertebrates with branches leading to human and to mouse marked as orange line.... Panels A and B show the assignments of new genes to branches on the phylogeny of human and mouse, respectively. The young genes, those primate- and rodent-specific, are marked with pink boxes. The notation with two numbers separated by a colon indicates the branch assignment and number of genes occurring in the given branch. For example, in the human lineage represented in panel A, in branch 12: 389 genes originated after the split of humans and chimps.... doi:10.1371/journal.pbio.1000494.g001 |
This team begins by identifying 1,828 human genes that are primate specific, and 3,111 mouse genes that are rodent-specific, calling them all "young genes". They then divide the young genes into three sub-categories, "DNA-level duplicates, RNA-level duplicates (retrogenes), and de novo genes." We note that genes in the first two sub-categories, the duplicates, come from pre-existing sequences. Thus, any programmatic content in these genes is not "new".
What about the de novo ones? According to the report, these genes have no annotated paralogs and no matches in existing databases. Darwinian theory gives no account of them. Such an account would show how their programmatic content was composed, step-by-step. Instead, they appear to have come, virtually finished, from nowhere.
Genes that are older than the features they encode and genes that come from nowhere, both are confounding for darwinism, because both acquired their programmatic content without the apparent benefit of mutation-and-selection. In cosmic ancestry, however, all genes are very old. The ones that seem to come from nowhere are merely being noticed for the first time.
We sincerely invite any pointers to counterexamples: If darwinian mutation-and-selection composes new genetic programs, where's the evidence?
 Zhang YE, Vibranovski MD, Landback P, Marais GAB, Long M, "Chromosomal Redistribution of Male-Biased Genes in Mammalian Evolution with Two Bursts of Gene Gain on the X Chromosome" [html], PLoS Biol 8(10): e1000494. doi:10.1371/journal.pbio.1000494, 5 Oct 2010. Zhang YE, Vibranovski MD, Landback P, Marais GAB, Long M, "Chromosomal Redistribution of Male-Biased Genes in Mammalian Evolution with Two Bursts of Gene Gain on the X Chromosome" [html], PLoS Biol 8(10): e1000494. doi:10.1371/journal.pbio.1000494, 5 Oct 2010.
 Metazoan Genes Older Than Metazoa? is a related local webpage. Metazoan Genes Older Than Metazoa? is a related local webpage.
 Three New Human Genes has become our main webpage about de novo genes. Three New Human Genes has become our main webpage about de novo genes.
 New genetic programs in Darwinism and strong panspermia is a related local webpage. New genetic programs in Darwinism and strong panspermia is a related local webpage.
 Chromosomal Redistribution... is the subject of our query to a coauthor of this study. He or she replies, but not for posting. No evidence is cited, and our quest for the missing evidence remains unanswered. Chromosomal Redistribution... is the subject of our query to a coauthor of this study. He or she replies, but not for posting. No evidence is cited, and our quest for the missing evidence remains unanswered.
 22 October 2010 22 October 2010
Gliese 581g may not exist. It was the Earthlike extrasolar planet whose discovery by astronomers from Universty of California, Santa Cruz, was reported last month. Now a rival team from the University of Geneva, examining additional data, see no evidence for the planet. Future observations should settle the issue.
 Richard A. Kerr, "First Goldilocks Exoplanet May Not Exist" [summary], doi:10.1126/science.330.6003.433, p433 v330, Science, 22 Oct 2010. Richard A. Kerr, "First Goldilocks Exoplanet May Not Exist" [summary], doi:10.1126/science.330.6003.433, p433 v330, Science, 22 Oct 2010.
 30 Sep 2010: Chances for life on this planet are 100 percent — our What'sNEW article about the reported discovery. 30 Sep 2010: Chances for life on this planet are 100 percent — our What'sNEW article about the reported discovery.
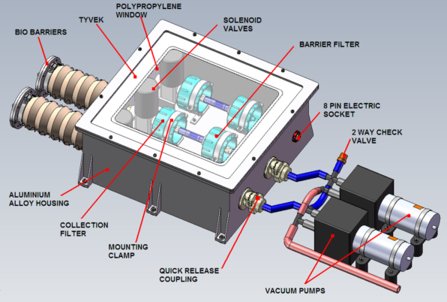
 6 October 2010 6 October 2010
Microbes in the high atmosphere will be sought by CASS-E, the University of Cranfield Astrobiological Stratospheric Sampling Experiment. A stratospheric balloon, carrying equipment (pictured) for collecting air samples at altitudes of 25 - 35 kilometers, is scheduled for launch from northern Sweden, this month. Strict precautions against contamination are a high priority, and panspermia is one of the topics that the students, who lead this project, will consider.
 Cranfield Astrobiological Stratospheric Sampling Experiment, homepage with links. Cranfield Astrobiological Stratospheric Sampling Experiment, homepage with links.
 An Atmospheric Test of Cometary Panspermia is a local webpage about an Indian balloon experiment, with links about subsequent testing. An Atmospheric Test of Cometary Panspermia is a local webpage about an Indian balloon experiment, with links about subsequent testing.
 Thanks, Nicholas Zumbulyadis. Thanks, Nicholas Zumbulyadis.
 1 October 2010 1 October 2010
|
GTAs produced by Reugeria mobilis |
Their only function seems to be transferring genes. Eugene Koonin, an evolutionary biologist at the National Institutes of Health in Bethesda, Maryland, is talking about gene transfer agents (GTAs). These are virus-like particles that contain strands of their host's genome inside a protein coat. When they emerge, a few host genes go with them. Thus they can efficiently shuttle genes between unrelated bacteria. "These particles... insert DNA into bacterial genomes so frequently that gene transfer in the ocean may occur 1,000 to 100 million times more often than previously thought. This suggests that GTAs have had a powerful role in evolution."
 Lauren D. McDaniel et al., "High Frequency of Horizontal Gene Transfer in the Oceans" [abstract], doi:10.1126/science.1192243, p50 v330, Science, 1 Oct 2010. Lauren D. McDaniel et al., "High Frequency of Horizontal Gene Transfer in the Oceans" [abstract], doi:10.1126/science.1192243, p50 v330, Science, 1 Oct 2010.
 Virus-like particles speed bacterial evolution, doi:10.1038/news.2010.507, by Amy Maxmen, Nature News, 30 Sep 2010. Virus-like particles speed bacterial evolution, doi:10.1038/news.2010.507, by Amy Maxmen, Nature News, 30 Sep 2010.
 Viruses and Other Gene Transfer Mechanisms is the main related local webpage — What'sNEW about HGT Viruses and Other Gene Transfer Mechanisms is the main related local webpage — What'sNEW about HGT  | |
 Thanks, Hans-Peter Wheeler. Thanks, Hans-Peter Wheeler.
 30 September 2010 30 September 2010
 Astronomers have found an extrasolar planet, Gliese 581g, considered the likeliest one to harbor life among the ~500 detected so far. Its orbit is smaller than Mercury's, but the star it circles, Gliese 581, is a red dwarf only 1% as bright as our sun. (The illustration shows that the orbit of the star's outermost planet, designated Gliese 581f, is slightly larger than Venus's.) Gliese 581g is tidally locked, so it has hot and cold hemispheres with a meridian of permanent "sunset" separating them. Liquid water could well persist within the temperature ranges on the planet. Co-discoverer Steven Vogt even says, Chances for life on this planet are 100 percent. Wow.
Astronomers have found an extrasolar planet, Gliese 581g, considered the likeliest one to harbor life among the ~500 detected so far. Its orbit is smaller than Mercury's, but the star it circles, Gliese 581, is a red dwarf only 1% as bright as our sun. (The illustration shows that the orbit of the star's outermost planet, designated Gliese 581f, is slightly larger than Venus's.) Gliese 581g is tidally locked, so it has hot and cold hemispheres with a meridian of permanent "sunset" separating them. Liquid water could well persist within the temperature ranges on the planet. Co-discoverer Steven Vogt even says, Chances for life on this planet are 100 percent. Wow.

 Astronomers Find Most Earth-like Planet to Date by Phil Berardelli, Science Now, 29 Sep 2010. Astronomers Find Most Earth-like Planet to Date by Phil Berardelli, Science Now, 29 Sep 2010.
 Could 'Goldilocks' planet be just right for life? by Seth Borenstein, Associated Press, posted on Yahoo!News, 29 Sep 2010. Could 'Goldilocks' planet be just right for life? by Seth Borenstein, Associated Press, posted on Yahoo!News, 29 Sep 2010.
 Newly Discovered Planet May Be First Truly Habitable Exoplanet, Press Release 10-172, National Science Foundation, 29 Sep 2010. Newly Discovered Planet May Be First Truly Habitable Exoplanet, Press Release 10-172, National Science Foundation, 29 Sep 2010.
 Life on Europa, Other Moons, Other Planets? has links about possible life on other worlds. Life on Europa, Other Moons, Other Planets? has links about possible life on other worlds.
 Thanks, Ellen Klyce (pictured). Thanks, Ellen Klyce (pictured).
 Gliese 581g may not exist — What'sNEW, 22 Oct 2010. Gliese 581g may not exist — What'sNEW, 22 Oct 2010.
 28 September 2010 28 September 2010
"...In plant-parasitic nematodes, a whole set of genes encoding proteins involved in the plant cell wall degradation was most likely acquired by LGT of bacterial origin. ...Selective advantage associated with transfer of these genes probably has driven their duplications and facilitated fixation in the different populations and species of plant-parasitic nematodes. Far from being negligible, these LGT events certainly have radically remolded evolutionary trends in recipient organisms, and similar roles in other animals can be expected to be discovered."
 Etienne G. J. Danchin et al., "Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes" [Open Access abstract], doi:10.1073/pnas.100848610, Proc. Natl. Acad. Sci. USA, online 27 Sep 2010. Etienne G. J. Danchin et al., "Multiple lateral gene transfers and duplications have promoted plant parasitism ability in nematodes" [Open Access abstract], doi:10.1073/pnas.100848610, Proc. Natl. Acad. Sci. USA, online 27 Sep 2010.
 Viruses and Other Gene Transfer Mechanisms is the main local webpage about LGT (or HGT) — What'sNEW about HGT Viruses and Other Gene Transfer Mechanisms is the main local webpage about LGT (or HGT) — What'sNEW about HGT  | |
 24 September 2010 24 September 2010
Seeking Signs of Life
9AM–4PM, Thursday, 14 Oct 2010
Lockheed Martin Global Vision Center
2121 Crystal Drive | Arlington, VA 22202 |
A one-day symposium "celebrating 50 Years of Exobiology and Astrobiology at NASA," featuring keynote speakers James Lovelock and Lynn Margulis, is open to the public. We regret that we cannot be there, but we think the signs of life beyond Earth seen by Richard Hoover and others, last described here on 15 Aug, should be included. We hope someone will mention them!
 First Announcement (PDF) of the Seeking Signs of Life symposium, with program and instructions for RSVP (required). First Announcement (PDF) of the Seeking Signs of Life symposium, with program and instructions for RSVP (required).
 More Evidence for Indigenous Microfossils in Carbonaceous Meteorites is the latest webpage about Hoover's evidence, posted 15 Aug 2010. More Evidence for Indigenous Microfossils in Carbonaceous Meteorites is the latest webpage about Hoover's evidence, posted 15 Aug 2010.
 Gaia is a related local webpage with more about James Lovelock. Gaia is a related local webpage with more about James Lovelock.
 Why Sexual Reproduction? is a local webpage that mentions Lynn Margulis. Why Sexual Reproduction? is a local webpage that mentions Lynn Margulis.
 20 September 2010 20 September 2010
Eukaryotic genes come from archaebacteria or eubacteria... or not. This is our micro-synopsis of a report by two leading geneticists from the British Isles who compared the genes of yeast and six other eukaryotes to those of prokaryotes. They found that 2,460 of 6,704 eukaryotic genes have prokaryotic homologs (see 3 examples below). Most of these homologs are found in eubacteria, but the ones from archaea are more important, they believe.
The two biologists mention a possibility: "These genes could be found in the yeast genome as a result of more recent lateral gene transfer (LGT), rather than being a relict of mitochondrial endosymbiosis." And they conclude, "Only half of eukaryotic genes have an identifiable prokaryotic homolog.... It is remarkable that some of the original partners' contributions might have persisted for > 1 billion years of evolution.... Yeast metabolism, and presumably eukaryotic metabolism in general, is a complex tapestry of prokaryotic threads and eukaryotic innovations."
Many eukaryotic genes have no known predecessors or homologs anywhere. This situation does not support the darwinian scheme, whereunder genes are composed gradually, by trial and error. Genes composed that way would leave a descending trail of identifiable precursors, but there seems to be no such trail. Instead, the genes are either present or absent, as observed here. That the innovations were composed by the darwinian method is supported only "presumably". Meanwhile, this research supports our hypothesis that all genes are very old "threads" that must be acquired, either vertically... or otherwise.
 James A. Cotton and James O. McInerney, "Eukaryotic genes of archaebacterial origin are more important than the more numerous eubacterial genes, irrespective of function" [abstract], doi:10.1073/pnas.1000265107, Proc. Natl. Acad. Sci. USA, online 17 Sep 2010. James A. Cotton and James O. McInerney, "Eukaryotic genes of archaebacterial origin are more important than the more numerous eubacterial genes, irrespective of function" [abstract], doi:10.1073/pnas.1000265107, Proc. Natl. Acad. Sci. USA, online 17 Sep 2010.
 New genetic programs in Darwinism and strong panspermia proposes a test for the provenance of "eukaryotic innovations." The original proposal contemplates mammals only, but the method is unrestricted. New genetic programs in Darwinism and strong panspermia proposes a test for the provenance of "eukaryotic innovations." The original proposal contemplates mammals only, but the method is unrestricted.
 13 September 2010 13 September 2010
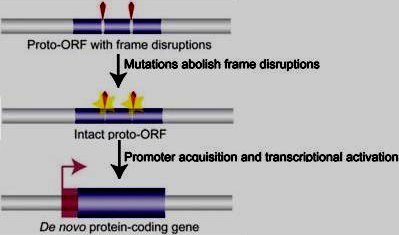
Origins, evolution, and phenotypic impact of new genes is the title of a review from the Center for Integrative Genomics at the University of Lausanne, Switzerland. In 14 pages with 127 references, author Henrik Kaessmann concludes, "Therefore, as had long been surmised (Ohno 1970), it is now beyond doubt that new genes have significantly contributed to organismal evolution." We have read the article closely to ask if the evidence in it supports strict darwinism or cosmic ancestry. In the former, new genes are invented stepwise, by trial and error, in a genetically closed system. In cosmic ancestry, new genes are only "new" in a local sense; the darwinian method cannot compose them from scratch, so they must be supplied, preexisting, in a genetically open system. Once supplied, genetic software management systems can assemble major pieces, and optimize them within limited ranges (and darwinian natural selection is needed for these processes.)
|
Figure 3. Origin of protein-coding genes from scratch. New coding regions may emerge de novo from noncoding genomic sequences. First, proto-open reading frames (proto-ORFs; thin blue bars) acquire mutations (point substitutions, insertions/deletions; yellow stars) that remove, bit by bit, frame-disrupting nucleotides (red wedges). Transcriptional activation of ORFs (through acquisition of promoters located in the 5' flanking region) encoding proteins with potentially useful functions may allow for the evolution of novel protein-coding genes. (Large blue box) Functional exon, (pink right-angled arrow) TSS, (transparent pink box) untranslated 5' sequence. Note that the transcriptional activation step may, alternatively, also precede the formation of complete functionally relevant ORFs. Figure and caption from Kaessmann, 2010. |
Our analysis begins with Kaessmann's summary of the ways new genes have originated —
1 "...new genes have arisen from copies of old ones...."
2 "...protein and RNA genes were composed from scratch...."
3 "...protein-coding genes metamorphosed into RNA genes...."
4 "...parasitic genome sequences were domesticated...."
5 "...the resulting components also readily mixed to yield new chimeric genes...."
We note that methods 1, 3 and 4 describe genes whose sequences are preexisting, needing only to be activated and optimized within limited ranges. Method 5 describes the assembly of genes from major pieces. Only method 2 looks like it would support strict darwinism. Let's look at it more closely.
The section titled "Gene origination from scratch" states, "In other words, new genes arise from previously nonfunctional genomic sequence, unrelated to any preexisting genic material (Fig. 3)." Already, new genes are seen come from pre-existing material, whose "origin" is unexplained. This section continues:
 "The de novo origin of entire protein-coding genes was long considered to be highly unlikely. For instance... François Jacob noted in an influential essay that the 'probability that a functional protein would appear de novo by random association of amino acids is practically zero' and that therefore the 'creation of entirely new nucleotide sequence could not be of any importance in the production of new information' (Jacob 1977)." "The de novo origin of entire protein-coding genes was long considered to be highly unlikely. For instance... François Jacob noted in an influential essay that the 'probability that a functional protein would appear de novo by random association of amino acids is practically zero' and that therefore the 'creation of entirely new nucleotide sequence could not be of any importance in the production of new information' (Jacob 1977)."
 "In spite of these notions, recent work has uncovered a number of new protein-coding genes that apparently arose from previously noncoding (and nonrepetitive) DNA sequences. Probably the first such case described in the literature is presented by the morpheus gene family that emerged in an Old World primate ancestor (Johnson et al. 2001). Although the details regarding the emergence of the original coding sequence remain unclear, the lack of any corresponding orthologous sequences outside of Old World primates suggest a de novo origin for this gene family...." Stepwise composition gets no support here. The sequence has no identifiable source. "In spite of these notions, recent work has uncovered a number of new protein-coding genes that apparently arose from previously noncoding (and nonrepetitive) DNA sequences. Probably the first such case described in the literature is presented by the morpheus gene family that emerged in an Old World primate ancestor (Johnson et al. 2001). Although the details regarding the emergence of the original coding sequence remain unclear, the lack of any corresponding orthologous sequences outside of Old World primates suggest a de novo origin for this gene family...." Stepwise composition gets no support here. The sequence has no identifiable source.
 "Other studies have followed suit and have provided a more detailed picture of de novo gene origination. For example, 14 de novo-originated genes have been identified in Drosophila (Levine et al. 2006; Zhou et al. 2008)...." We analysed those reports when they were current, finding poor evidence for stepwise composition. Links are below. "Other studies have followed suit and have provided a more detailed picture of de novo gene origination. For example, 14 de novo-originated genes have been identified in Drosophila (Levine et al. 2006; Zhou et al. 2008)...." We analysed those reports when they were current, finding poor evidence for stepwise composition. Links are below.
 "For example, Knowles and McLysaght (2009) recently identified three genes that seem to have arisen from scratch on the human lineage. Detailed analyses of these human-specific genes, which involved comparisons with corresponding noncoding sequences from closely related primate relatives, revealed that a few mutational events after the separation of the human and chimpanzee lineages abolished 'disabling' nucleotides in the ancestral open reading frame precursors (Fig. 3 [above]), allowing relatively long coding sequences to emanate in humans. Importantly, the functionality of these new human genes is supported by evidence for translation of their coding sequences...." The figure shows a precursor gene apparently 95% composed, needing only 2 point mutations to restore the ORF. Deeper analyis is available on our webpage "Three New Human Genes," posted a year ago, linked below. "For example, Knowles and McLysaght (2009) recently identified three genes that seem to have arisen from scratch on the human lineage. Detailed analyses of these human-specific genes, which involved comparisons with corresponding noncoding sequences from closely related primate relatives, revealed that a few mutational events after the separation of the human and chimpanzee lineages abolished 'disabling' nucleotides in the ancestral open reading frame precursors (Fig. 3 [above]), allowing relatively long coding sequences to emanate in humans. Importantly, the functionality of these new human genes is supported by evidence for translation of their coding sequences...." The figure shows a precursor gene apparently 95% composed, needing only 2 point mutations to restore the ORF. Deeper analyis is available on our webpage "Three New Human Genes," posted a year ago, linked below.
We think this review of the origins of new genes, purporting to sustain the darwinian account of them, fails. We think a similar failure is quite common in darwinists' writings. We wish that instances of this failure were noticed and discussed openly. Moreover, we think the cited evidence, and lots more, points instead to genes that are supplied, preexisting, whole or in major pieces — which darwinism cannot explain, but which cosmic ancestry requires.
 Henrik Kaessmann, "Origins, evolution, and phenotypic impact of new genes" [abstract], doi:10.1101/gr.101386.109, Genome Research, online 22 Jul 2010. Henrik Kaessmann, "Origins, evolution, and phenotypic impact of new genes" [abstract], doi:10.1101/gr.101386.109, Genome Research, online 22 Jul 2010.
 Novel genes derived from noncoding DNA is our article about the Drosophila study by Levine et al., posted 15 Jun 2006. Novel genes derived from noncoding DNA is our article about the Drosophila study by Levine et al., posted 15 Jun 2006.
 The origin of new genes is our article about the study of fruitflies by Zhou et al., posted 17 Aug 2008. The origin of new genes is our article about the study of fruitflies by Zhou et al., posted 17 Aug 2008.
 Three New Human Genes is the local webpage discussing Knowles and McLysaght, posted 4 Sep 2009. Three New Human Genes is the local webpage discussing Knowles and McLysaght, posted 4 Sep 2009.
 A Wordcount for Comparison is a related local webpage. A Wordcount for Comparison is a related local webpage.
 New genetic programs in Darwinism and strong panspermia is a related local webpage. New genetic programs in Darwinism and strong panspermia is a related local webpage.
 Viruses and Other Gene Transfer Mechanisms is the main local webpage about how genes can be supplied — What'sNEW about HGT Viruses and Other Gene Transfer Mechanisms is the main local webpage about how genes can be supplied — What'sNEW about HGT  | |
 9 September 2010 9 September 2010
Evolutionary origin of HPG? Eight marine biologists and biochemists from Japan and one from the US claim to have discovered the likely origin of the hypothalamic-pituitary-gonadal (HPG) axis, an endocrine system that is "remarkably conserved" among jawed vertebrates. They note, Such an evolutionary innovation is one of the key elements leading to physiological divergence, including reproduction, growth, metabolism, stress, and osmoregulation in subsequent evolution of jawed vertebrates.... The data supporting the conclusion follow a phylogenetic analysis (illustrated) of 80 deduced amino-acid sequences of the two HPG protein subunits found in hagfish (highlighted) that were the focus of the study. [Click to see full image and legend in a new window.]
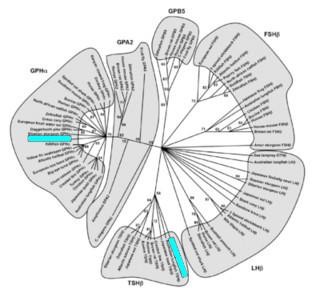 The thesis sentences of the article read, We hypothesize that the identity of a single, functional GPH... in hagfish, provides critical evidence for the existence of a HPG system in the most basal vertebrate. Furthermore, we propose that this HPG system likely evolved from an ancestral, prevertebrate exclusively neuroendocrine mechanism by gradual emergence of components of a previously undescribed control level, the pituitary, which is not present in the Protochordates.
The thesis sentences of the article read, We hypothesize that the identity of a single, functional GPH... in hagfish, provides critical evidence for the existence of a HPG system in the most basal vertebrate. Furthermore, we propose that this HPG system likely evolved from an ancestral, prevertebrate exclusively neuroendocrine mechanism by gradual emergence of components of a previously undescribed control level, the pituitary, which is not present in the Protochordates.
The research is thoroughgoing and admirable, but, having studied the report closely, we note that it has nothing to say about the "origin" of this endocrine system. "Origin" means source, inception, starting point, or beginning. Instead, this team reports, With the discovery of... [identifiable] homologs not only in other vertebrates but invertebrates—including fly, nematode, and sea urchin... it is proposed that an ancestral glycoprotein (GP) existed before the divergence of vertebrates and invertebrates.... An ancestral GP likely existed in the common ancestor of protostomes and deuterostomes, and diverged then... in both lineages.
The precursor genes apparently already existed among deep ancestral species, with nothing similar found beyond (earlier than) that. That they "likely evolved," as required in standard darwinism, is a complete, obvious supposition. The evidence shows only that they were absent until they were present. Once present, their history can be estimated, but their origin is as unexplained as ever. This misrepresentation — that an origin has been revealed when it hasn't — is common, in our experience. We welcome pointers to counterexamples.
Meanwhile, genes that are older than fossil evidence would suggest, and genes that appear abruptly without identifiable precursors, confirm basic predictions of cosmic ancestry. After they are acquired, genes may duplicate and diverge within limited ranges, as this research observes for HPG.
 Katsuhisa Uchida et al., "Evolutionary origin of a functional gonadotropin in the pituitary of the most primitive vertebrate, hagfish" [Open Access abstract], doi:10.1073/pnas.1002208107, p15832-15837 v107, Proc. Natl. Acad. Sci. USA, 7 Sep 2010. Katsuhisa Uchida et al., "Evolutionary origin of a functional gonadotropin in the pituitary of the most primitive vertebrate, hagfish" [Open Access abstract], doi:10.1073/pnas.1002208107, p15832-15837 v107, Proc. Natl. Acad. Sci. USA, 7 Sep 2010.
 Metazoan Genes Older Than Metazoa? is a related local webpage. Metazoan Genes Older Than Metazoa? is a related local webpage.
 Testing Darwinism versus Cosmic Ancestry is a related local webpage. Testing Darwinism versus Cosmic Ancestry is a related local webpage.
|

 Astronomers have found an extrasolar planet, Gliese 581g, considered the likeliest one to harbor life among the ~500 detected so far. Its orbit is smaller than Mercury's, but the star it circles, Gliese 581, is a red dwarf only 1% as bright as our sun. (The illustration shows that the orbit of the star's outermost planet, designated Gliese 581f, is slightly larger than Venus's.) Gliese 581g is tidally locked, so it has hot and cold hemispheres with a meridian of permanent "sunset" separating them. Liquid water could well persist within the temperature ranges on the planet. Co-discoverer Steven Vogt even says, Chances for life on this planet are 100 percent. Wow.
Astronomers have found an extrasolar planet, Gliese 581g, considered the likeliest one to harbor life among the ~500 detected so far. Its orbit is smaller than Mercury's, but the star it circles, Gliese 581, is a red dwarf only 1% as bright as our sun. (The illustration shows that the orbit of the star's outermost planet, designated Gliese 581f, is slightly larger than Venus's.) Gliese 581g is tidally locked, so it has hot and cold hemispheres with a meridian of permanent "sunset" separating them. Liquid water could well persist within the temperature ranges on the planet. Co-discoverer Steven Vogt even says, Chances for life on this planet are 100 percent. Wow.
 "The diverse coding potential of CroV includes predicted translation factors, DNA repair enzymes such as DNA mismatch repair protein MutS and two photolyases, multiple ubiquitin pathway components, four intein elements, and 22 tRNAs. Many genes including isoleucyl-tRNA synthetase, eIF-2γ, and an Elp3-like histone acetyltransferase are usually not found in viruses. We also discovered a 38-kb genomic region of putative bacterial origin, which encodes several predicted carbohydrate metabolizing enzymes, including an entire pathway for the biosynthesis of 3-deoxy-d-manno-octulosonate, a key component of the outer membrane in Gram-negative bacteria."
"The diverse coding potential of CroV includes predicted translation factors, DNA repair enzymes such as DNA mismatch repair protein MutS and two photolyases, multiple ubiquitin pathway components, four intein elements, and 22 tRNAs. Many genes including isoleucyl-tRNA synthetase, eIF-2γ, and an Elp3-like histone acetyltransferase are usually not found in viruses. We also discovered a 38-kb genomic region of putative bacterial origin, which encodes several predicted carbohydrate metabolizing enzymes, including an entire pathway for the biosynthesis of 3-deoxy-d-manno-octulosonate, a key component of the outer membrane in Gram-negative bacteria."